Table of Contents
To perform a Lack of Fit Test in R, first define the model of interest, then fit the model to the data, and use the anova() function to compare the full model to a reduced model. The reduced model should be a subset of the full model, and should not include any terms that are part of the lack of fit test. Finally, calculate the Lack of Fit Test statistic and compare it to the F critical value to determine if the lack of fit is significant.
A lack of fit test is used to determine whether or not a full offers a significantly better fit to a dataset than some reduced version of the model.
For example, suppose we would like to use number of hours studied to predict exam score for students at a certain college. We may decide to fit the following two regression models:
Full Model: Score = β0 + B1(hours) + B2(hours)2
Reduced Model: Score = β0 + B1(hours)
The following step-by-step example shows how to perform a lack of fit test in R to determine if the full model offers a significantly better fit than the reduced model.
Step 1: Create & Visualize a Dataset
First, we’ll use the following code to create a dataset that contains the number of hours studied and exam score received for 50 students:
#make this example reproducible set.seed(1) #create dataset df <- data.frame(hours = runif(50, 5, 15), score=50) df$score = df$score + df$hours^3/150 + df$hours*runif(50, 1, 2) #view first six rows of data head(df) hours score 1 7.655087 64.30191 2 8.721239 70.65430 3 10.728534 73.66114 4 14.082078 86.14630 5 7.016819 59.81595 6 13.983897 83.60510
Next, we’ll create a scatterplot to visualize the relationship between hours and score:
#load ggplot2 visualization package library(ggplot2) #create scatterplot ggplot(df, aes(x=hours, y=score)) + geom_point()
Step 2: Fit Two Different Models to the Dataset
Next, we’ll fit two different regression models to the dataset:
#fit full model full <- lm(score ~ poly(hours,2), data=df) #fit reduced model reduced <- lm(score ~ hours, data=df)
Step 3: Perform a Lack of Fit Test
Next, we’ll use the anova() command to perform a lack of fit test between the two models:
#lack of fit test
anova(full, reduced)
Analysis of Variance Table
Model 1: score ~ poly(hours, 2)
Model 2: score ~ hours
Res.Df RSS Df Sum of Sq F Pr(>F)
1 47 368.48
2 48 451.22 -1 -82.744 10.554 0.002144 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Step 4: Visualize the Final Model
Lastly, we can visualize the final model (the full model) relative to the original dataset:
ggplot(df, aes(x=hours, y=score)) +
geom_point() +
stat_smooth(method='lm', formula = y ~ poly(x,2), size = 1) +
xlab('Hours Studied') +
ylab('Score')
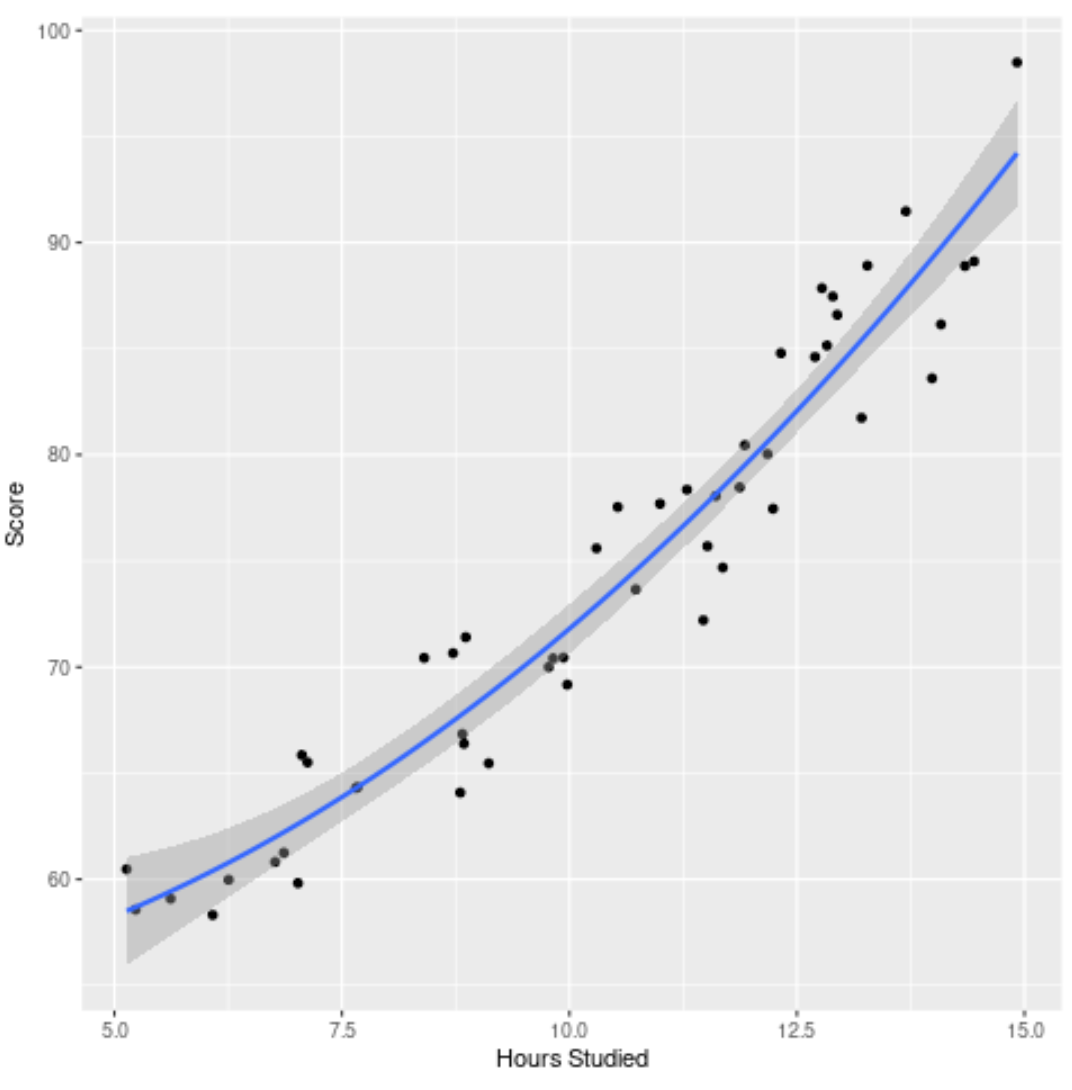
We can see that the curve of the model fits the data quite well.
