Table of Contents
Principal Components Regression (PCR) is a method of regression analysis that is used to reduce the number of predictor variables in a data set by using principal components to replace the original variables. This method can help to reduce the collinearity between variables and improve the model accuracy. To perform PCR in R, you must first create the model matrix, then calculate the principal components, and finally perform the regression. This can be done with the prcomp(), predict(), and lm() functions in R.
Given a set of p predictor variables and a response variable, multiple linear regression uses a method known as least squares to minimize the sum of squared residuals (RSS):
RSS = Σ(yi – ŷi)2
where:
- Σ: A greek symbol that means sum
- yi: The actual response value for the ith observation
- ŷi: The predicted response value based on the multiple linear regression model
However, when the predictor variables are highly correlated then multicollinearity can become a problem. This can cause the coefficient estimates of the model to be unreliable and have high variance.
One way to avoid this problem is to instead use principal components regression, which finds M linear combinations (known as “principal components”) of the original p predictors and then uses least squares to fit a linear regression model using the principal components as predictors.
This tutorial provides a step-by-step example of how to perform principal components regression in R.
Step 1: Load Necessary Packages
The easiest way to perform principal components regression in R is by using functions from the pls package.
#install pls package (if not already installed) install.packages("pls") load pls package library(pls)
Step 2: Fit PCR Model
For this example, we’ll use the built-in R dataset called mtcars which contains data about various types of cars:
#view first six rows of mtcars dataset
head(mtcars)
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
For this example we’ll fit a principal components regression (PCR) model using hp as the response variable and the following variables as the predictor variables:
- mpg
- disp
- drat
- wt
- qsec
The following code shows how to fit the PCR model to this data. Note the following arguments:
- scale=TRUE: This tells R that each of the predictor variables should be scaled to have a mean of 0 and a standard deviation of 1. This ensures that no predictor variable is overly influential in the model if it happens to be measured in different units.
- validation=”CV”: This tells R to use k-fold cross-validation to evaluate the performance of the model. Note that this uses k=10 folds by default. Also note that you can specify “LOOCV” instead to perform leave-one-out cross-validation.
#make this example reproducible set.seed(1) #fit PCR model model <- pcr(hp~mpg+disp+drat+wt+qsec, data=mtcars, scale=TRUE, validation="CV")
Step 3: Choose the Number of Principal Components
Once we’ve fit the model, we need to determine the number of principal components worth keeping.
The way to do so is by looking at the test root mean squared error (test RMSE) calculated by the k-fold cross-validation:
#view summary of model fitting
summary(model)
Data: X dimension: 32 5
Y dimension: 32 1
Fit method: svdpc
Number of components considered: 5
VALIDATION: RMSEP
Cross-validated using 10 random segments.
(Intercept) 1 comps 2 comps 3 comps 4 comps 5 comps
CV 69.66 44.56 35.64 35.83 36.23 36.67
adjCV 69.66 44.44 35.27 35.43 35.80 36.20
TRAINING: % variance explained
1 comps 2 comps 3 comps 4 comps 5 comps
X 69.83 89.35 95.88 98.96 100.00
hp 62.38 81.31 81.96 81.98 82.03
There are two tables of interest in the output:
1. VALIDATION: RMSEP
This table tells us the test RMSE calculated by the k-fold cross validation. We can see the following:
- If we only use the intercept term in the model, the test RMSE is 69.66.
- If we add in the first principal component, the test RMSE drops to 44.56.
- If we add in the second principal component, the test RMSE drops to 35.64.
We can see that adding additional principal components actually leads to an increase in test RMSE. Thus, it appears that it would be optimal to only use two principal components in the final model.
2. TRAINING: % variance explained
This table tells us the percentage of the variance in the response variable explained by the principal components. We can see the following:
- By using just the first principal component, we can explain 69.83% of the variation in the response variable.
- By adding in the second principal component, we can explain 89.35% of the variation in the response variable.
Note that we’ll always be able to explain more variance by using more principal components, but we can see that adding in more than two principal components doesn’t actually increase the percentage of explained variance by much.
We can also visualize the test RMSE (along with the test MSE and R-squared) based on the number of principal components by using the validationplot() function.
#visualize cross-validation plots
validationplot(model)
validationplot(model, val.type="MSEP")
validationplot(model, val.type="R2")
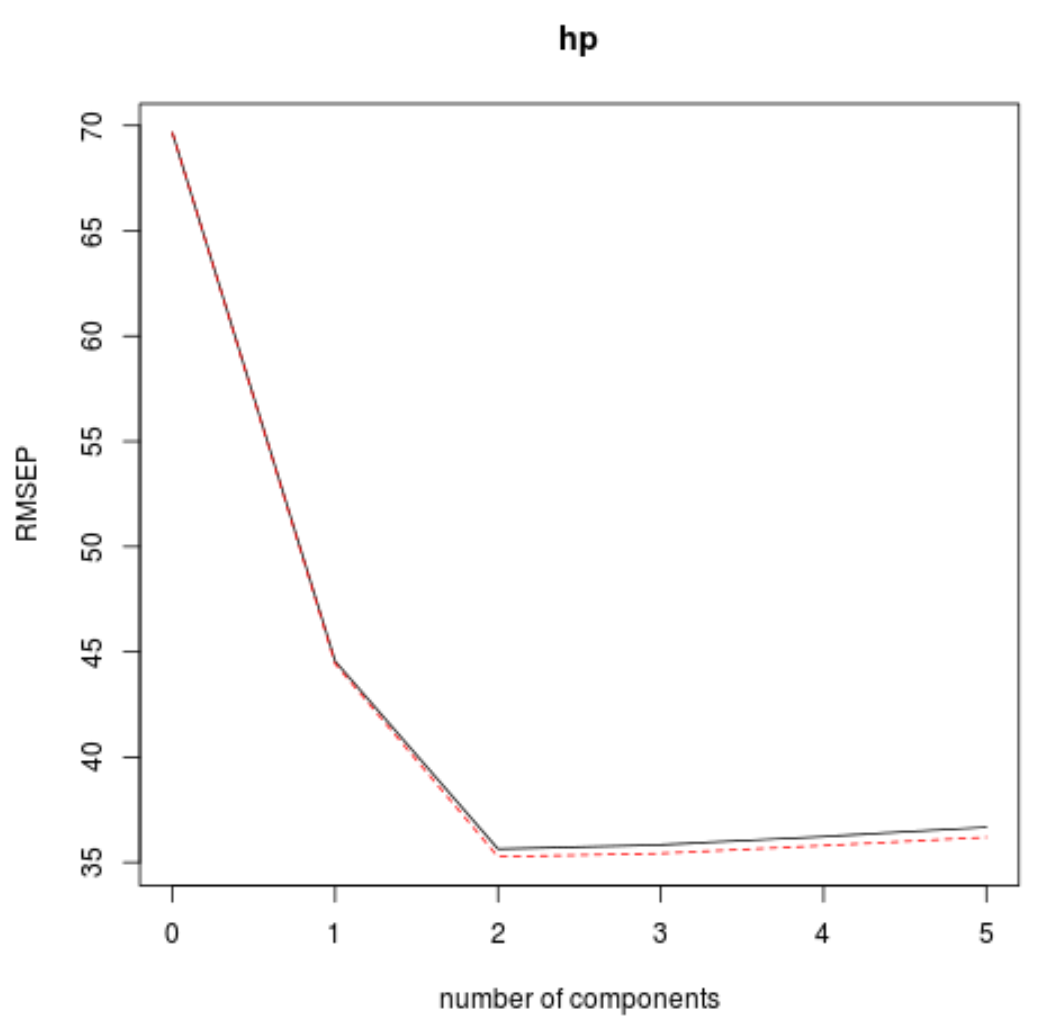
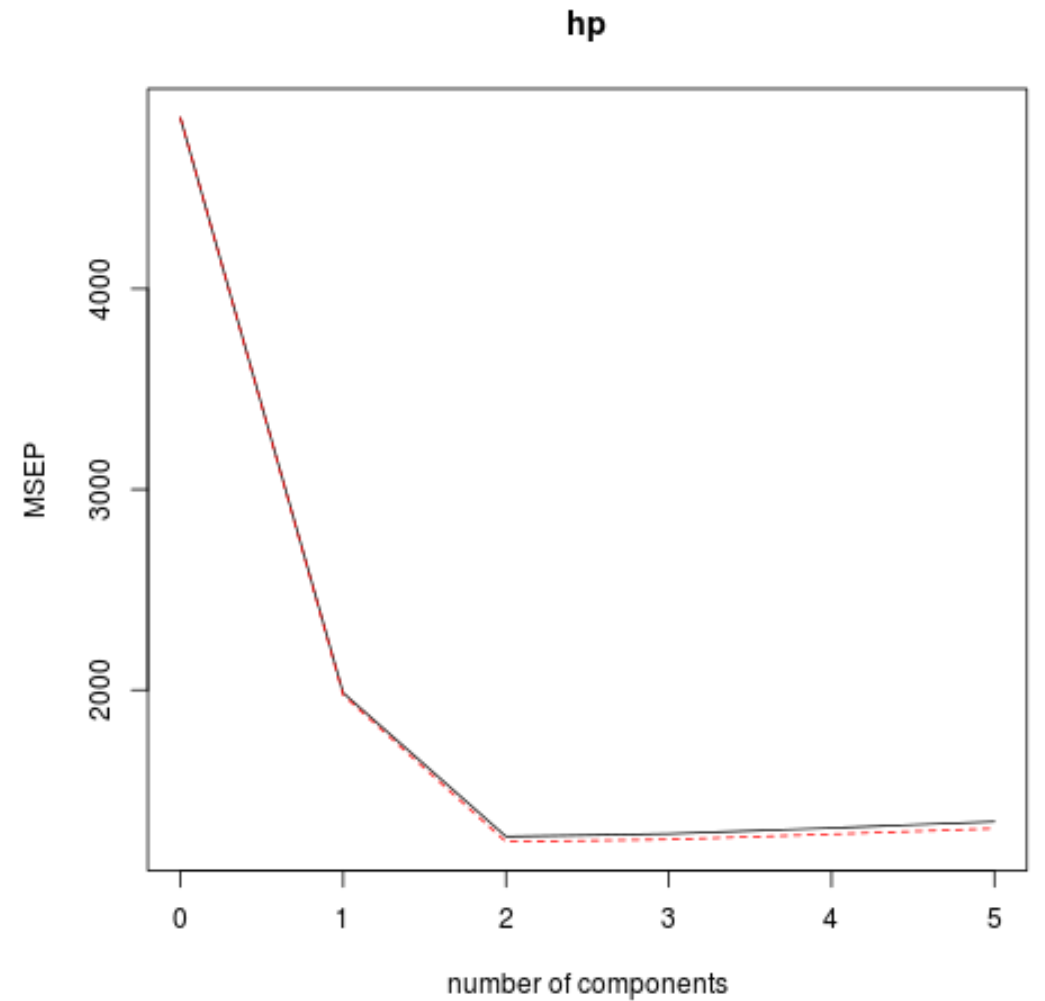
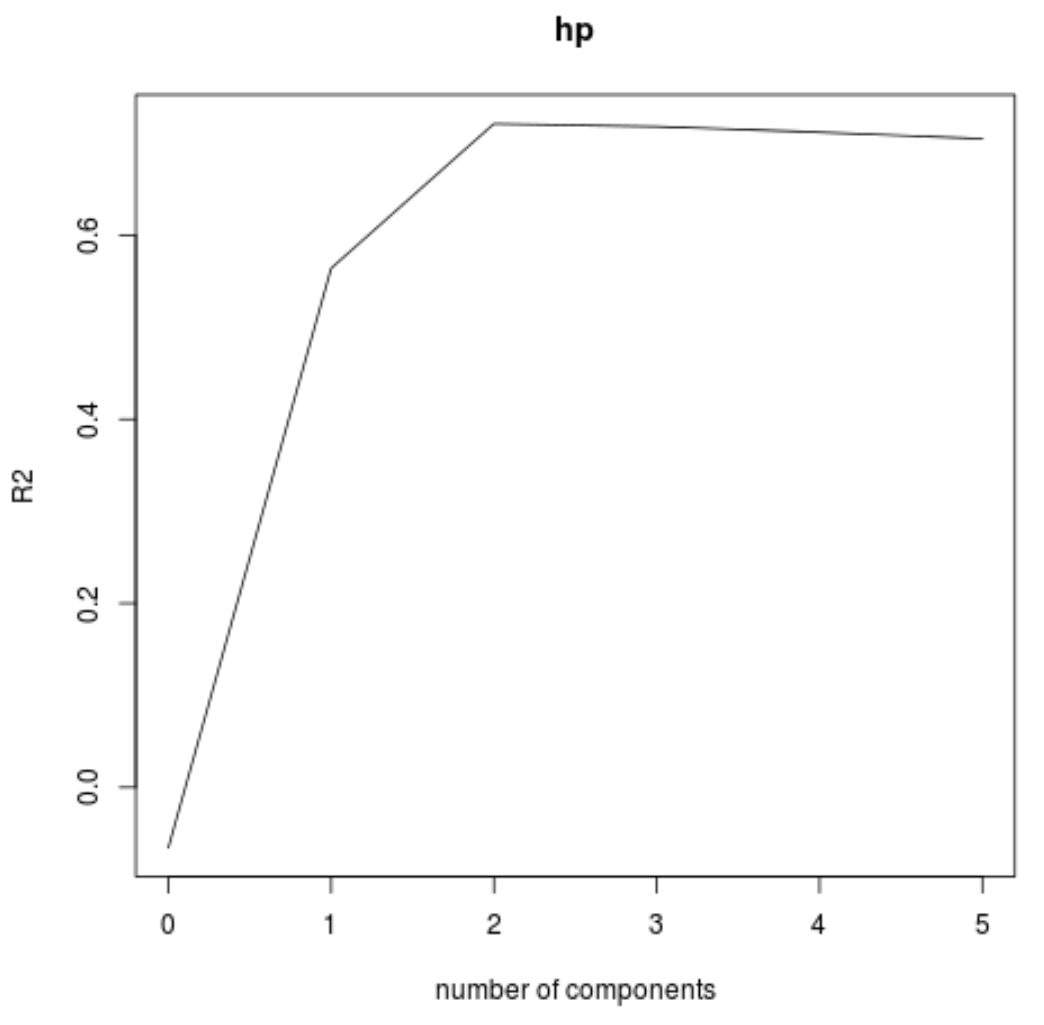
In each plot we can see that the model fit improves by adding in two principal components, yet it tends to get worse when we add more principal components.
Thus, the optimal model includes just the first two principal components.
Step 4: Use the Final Model to Make Predictions
We can use the final PCR model with two principal components to make predictions on new observations.
The following code shows how to split the original dataset into a training and testing set and use the PCR model with two principal components to make predictions on the testing set.
#define training and testing sets train <- mtcars[1:25, c("hp", "mpg", "disp", "drat", "wt", "qsec")] y_test <- mtcars[26:nrow(mtcars), c("hp")] test <- mtcars[26:nrow(mtcars), c("mpg", "disp", "drat", "wt", "qsec")] #use model to make predictions on a test set model <- pcr(hp~mpg+disp+drat+wt+qsec, data=train, scale=TRUE, validation="CV") pcr_pred <- predict(model, test, ncomp=2) #calculate RMSE sqrt(mean((pcr_pred - y_test)^2)) [1] 56.86549
We can see that the test RMSE turns out to be 56.86549. This is the average deviation between the predicted value for hp and the observed value for hp for the observations in the testing set.
The complete R code use in this example can be found here.
