Table of Contents
Bootstrapping is a statistical resampling technique used to estimate the variability and uncertainty of a population parameter by repeatedly sampling from a given dataset. In R, bootstrapping can be performed using the “boot” package, which allows for the creation of a large number of resampled datasets from the original data. These resampled datasets can then be used to calculate the desired statistic and generate a confidence interval for its true value. Some examples of bootstrapping in R include estimating the mean or median of a population, creating a confidence interval for a regression coefficient, or comparing the means of two groups. Bootstrapping is a useful tool in situations where traditional statistical methods may not be appropriate, such as when the underlying data is not normally distributed or when the sample size is small. It is a versatile and powerful technique that can provide valuable insights into the uncertainty of our data.
Perform Bootstrapping in R (With Examples)
Bootstrapping is a method that can be used to estimate the standard error of any statistic and produce a confidence interval for the statistic.
The basic process for bootstrapping is as follows:
- Take k repeated samples with replacement from a given dataset.
- For each sample, calculate the statistic you’re interested in.
- This results in k different estimates for a given statistic, which you can then use to calculate the standard error of the statistic and create a confidence interval for the statistic.
We can perform bootstrapping in R by using the following functions from the boot library:
1. Generate bootstrap samples.
boot(data, statistic, R, …)
where:
- data: A vector, matrix, or data frame
- statistic: A function that produces the statistic(s) to be bootstrapped
- R: Number of bootstrap replicates
2. Generate a bootstrapped confidence interval.
boot.ci(bootobject, conf, type)
where:
- bootobject: An object returned by the boot() function
- conf: The confidence interval to calculate. Default is 0.95
- type: Type of confidence interval to calculate. Options include “norm”, “basic”, “stud”, “perc”, “bca” and “all” – Default is “all”
The following examples show how to use these functions in practice.
Example 1: Bootstrap a Single Statistic
The following code shows how to calculate the standard error for the R-squared of a simple linear regression model:
set.seed(0) library(boot) #define function to calculate R-squared rsq_function <- function(formula, data, indices) { d <- data[indices,] #allows boot to select sample fit <- lm(formula, data=d) #fit regression modelreturn(summary(fit)$r.square) #return R-squared of model } #perform bootstrapping with 2000 replications reps <- boot(data=mtcars, statistic=rsq_function, R=2000, formula=mpg~disp) #view results of boostrapping reps ORDINARY NONPARAMETRIC BOOTSTRAP Call: boot(data = mtcars, statistic = rsq_function, R = 2000, formula = mpg ~ disp) Bootstrap Statistics : original bias std. error t1* 0.7183433 0.002164339 0.06513426
From the results we can see:
- The estimated R-squared for this regression model is 0.7183433.
- The standard error for this estimate is 0.06513426.
We can quickly view the distribution of the bootstrapped samples as well:
plot(reps)
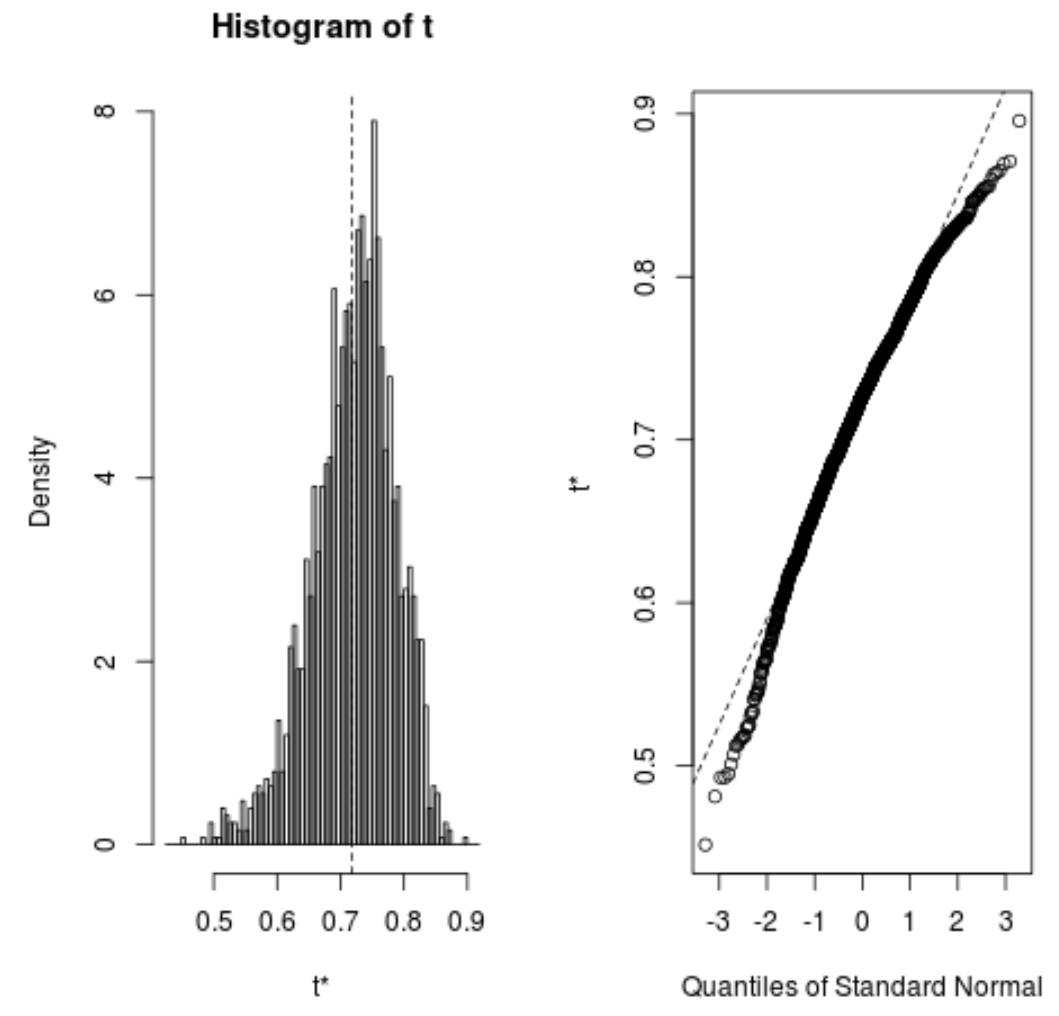
We can also use the following code to calculate the 95% confidence interval for the estimated R-squared of the model:
#calculate adjusted bootstrap percentile (BCa) interval boot.ci(reps, type="bca") CALL : boot.ci(boot.out = reps, type = "bca") Intervals : Level BCa 95% ( 0.5350, 0.8188 ) Calculations and Intervals on Original Scale
From the output we can see that the 95% bootstrapped confidence interval for the true R-squared values is (.5350, .8188).
Example 2: Bootstrap Multiple Statistics
The following code shows how to calculate the standard error for each coefficient in a multiple linear regression model:
set.seed(0) library(boot) #define function to calculate fitted regression coefficients coef_function <- function(formula, data, indices) { d <- data[indices,] #allows boot to select sample fit <- lm(formula, data=d) #fit regression modelreturn(coef(fit)) #return coefficient estimates of model } #perform bootstrapping with 2000 replications reps <- boot(data=mtcars, statistic=coef_function, R=2000, formula=mpg~disp) #view results of boostrapping reps ORDINARY NONPARAMETRIC BOOTSTRAP Call: boot(data = mtcars, statistic = coef_function, R = 2000, formula = mpg ~ disp) Bootstrap Statistics : original bias std. error t1* 29.59985476 -5.058601e-02 1.49354577 t2* -0.04121512 6.549384e-05 0.00527082
From the results we can see:
- The estimated coefficient for the intercept of the model is 29.59985476 and the standard error of this estimate is 1.49354577.
- The estimated coefficient for the predictor variable disp in the model is -0.04121512 and the standard error of this estimate is 0.00527082.
We can quickly view the distribution of the bootstrapped samples as well:
plot(reps, index=1) #intercept of model plot(reps, index=2) #disp predictor variable
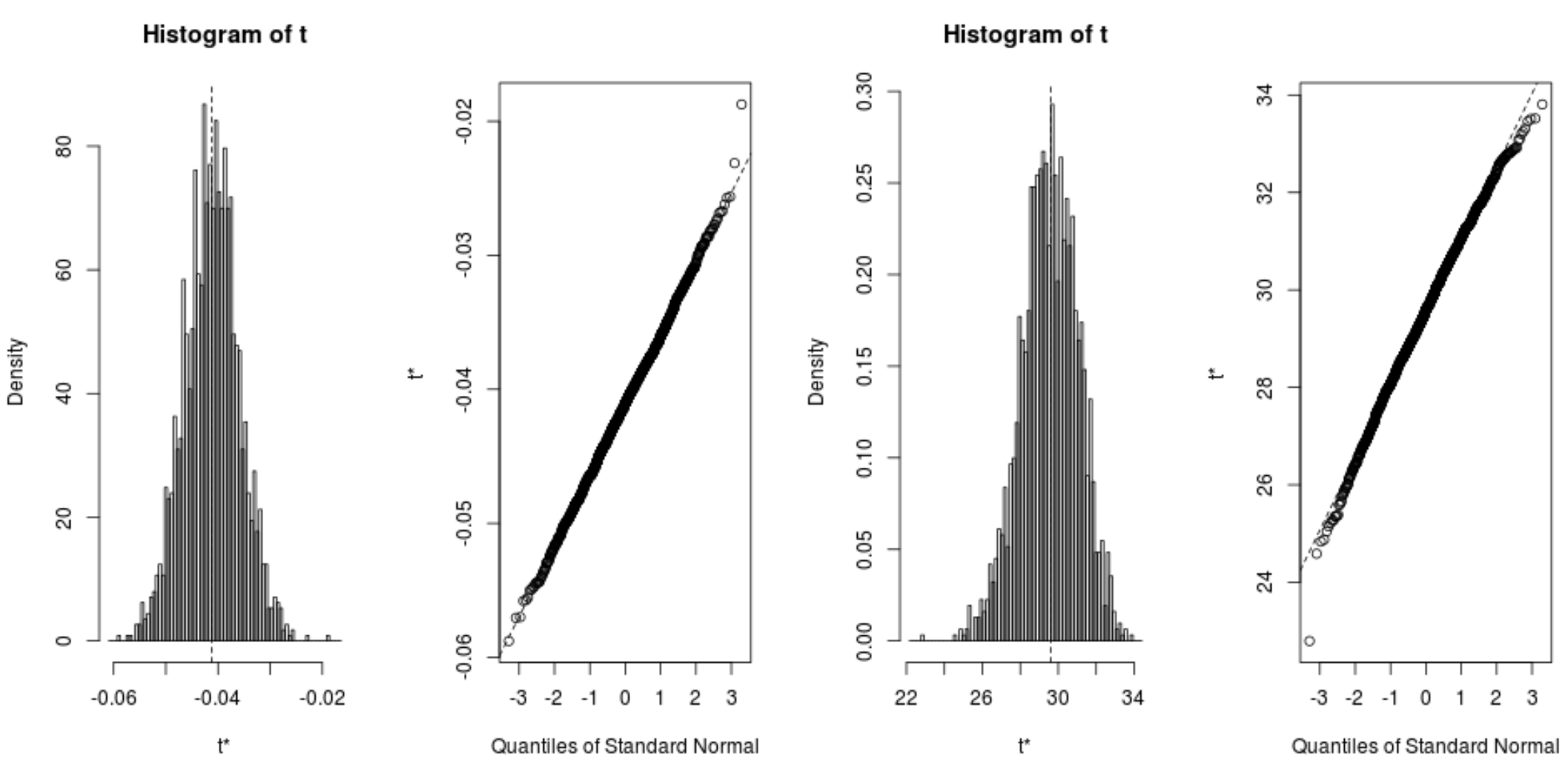
We can also use the following code to calculate the 95% confidence intervals for each coefficient:
#calculate adjusted bootstrap percentile (BCa) intervals boot.ci(reps, type="bca", index=1) #intercept of model boot.ci(reps, type="bca", index=2) #disp predictor variable CALL : boot.ci(boot.out = reps, type = "bca", index = 1) Intervals : Level BCa 95% (26.78, 32.66 ) Calculations and Intervals on Original Scale BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS Based on 2000 bootstrap replicates CALL : boot.ci(boot.out = reps, type = "bca", index = 2) Intervals : Level BCa 95% (-0.0520, -0.0312 ) Calculations and Intervals on Original Scale
From the output we can see that the 95% bootstrapped confidence intervals for the model coefficients are as follows:
- C.I. for intercept: (26.78, 32.66)
- C.I. for disp: (-.0520, -.0312)
Additional Resources
How to Perform Simple Linear Regression in R
How to Perform Multiple Linear Regression in R
Introduction to Confidence Intervals
