Table of Contents
Bootstrapping in R is a resampling technique used to estimate the sampling distribution of a statistic by drawing data from a sample with replacement. It is a useful tool for analyzing data when there is a limited amount of it available. To perform bootstrapping in R, you must first create a bootstrap function, then apply it to the data set to obtain the desired result. Examples of bootstrapping in R can be found in the R documentation for the boot package.
Bootstrapping is a method that can be used to estimate the standard error of any statistic and produce a confidence interval for the statistic.
The basic process for bootstrapping is as follows:
- Take k repeated samples with replacement from a given dataset.
- For each sample, calculate the statistic you’re interested in.
- This results in k different estimates for a given statistic, which you can then use to calculate the standard error of the statistic and create a confidence interval for the statistic.
We can perform bootstrapping in R by using the following functions from the boot library:
1. Generate bootstrap samples.
boot(data, statistic, R, …)
where:
- data: A vector, matrix, or data frame
- statistic: A function that produces the statistic(s) to be bootstrapped
- R: Number of bootstrap replicates
2. Generate a bootstrapped confidence interval.
boot.ci(bootobject, conf, type)
where:
- bootobject: An object returned by the boot() function
- conf: The confidence interval to calculate. Default is 0.95
- type: Type of confidence interval to calculate. Options include “norm”, “basic”, “stud”, “perc”, “bca” and “all” – Default is “all”
The following examples show how to use these functions in practice.
Example 1: Bootstrap a Single Statistic
The following code shows how to calculate the standard error for the R-squared of a simple linear regression model:
set.seed(0) library(boot) #define function to calculate R-squared rsq_function <- function(formula, data, indices) { d <- data[indices,] #allows boot to select sample fit <- lm(formula, data=d) #fit regression model return(summary(fit)$r.square) #return R-squared of model } #perform bootstrapping with 2000 replications reps <- boot(data=mtcars, statistic=rsq_function, R=2000, formula=mpg~disp) #view results of boostrapping reps ORDINARY NONPARAMETRIC BOOTSTRAP Call: boot(data = mtcars, statistic = rsq_function, R = 2000, formula = mpg ~ disp) Bootstrap Statistics : original bias std. error t1* 0.7183433 0.002164339 0.06513426
From the results we can see:
- The estimated R-squared for this regression model is 0.7183433.
- The standard error for this estimate is 0.06513426.
We can quickly view the distribution of the bootstrapped samples as well:
plot(reps)
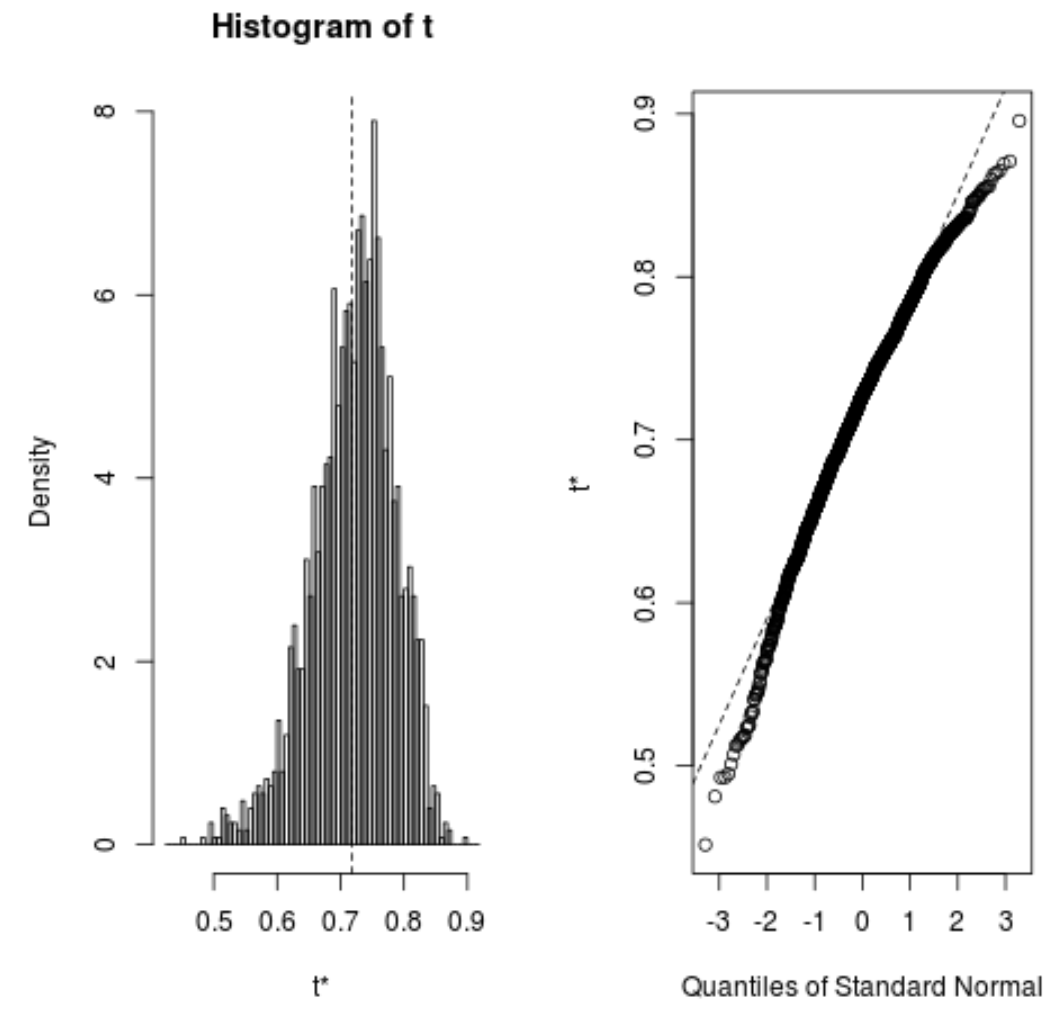
We can also use the following code to calculate the 95% confidence interval for the estimated R-squared of the model:
#calculate adjusted bootstrap percentile (BCa) interval boot.ci(reps, type="bca") CALL : boot.ci(boot.out = reps, type = "bca") Intervals : Level BCa 95% ( 0.5350, 0.8188 ) Calculations and Intervals on Original Scale
From the output we can see that the 95% bootstrapped confidence interval for the true R-squared values is (.5350, .8188).
Example 2: Bootstrap Multiple Statistics
The following code shows how to calculate the standard error for each coefficient in a multiple linear regression model:
set.seed(0) library(boot) #define function to calculate fitted regression coefficients coef_function <- function(formula, data, indices) { d <- data[indices,] #allows boot to select sample fit <- lm(formula, data=d) #fit regression model return(coef(fit)) #return coefficient estimates of model } #perform bootstrapping with 2000 replications reps <- boot(data=mtcars, statistic=coef_function, R=2000, formula=mpg~disp) #view results of boostrapping reps ORDINARY NONPARAMETRIC BOOTSTRAP Call: boot(data = mtcars, statistic = coef_function, R = 2000, formula = mpg ~ disp) Bootstrap Statistics : original bias std. error t1* 29.59985476 -5.058601e-02 1.49354577 t2* -0.04121512 6.549384e-05 0.00527082
From the results we can see:
- The estimated coefficient for the intercept of the model is 29.59985476 and the standard error of this estimate is 1.49354577.
- The estimated coefficient for the predictor variable disp in the model is -0.04121512 and the standard error of this estimate is 0.00527082.
We can quickly view the distribution of the bootstrapped samples as well:
plot(reps, index=1) #intercept of model plot(reps, index=2) #disp predictor variable
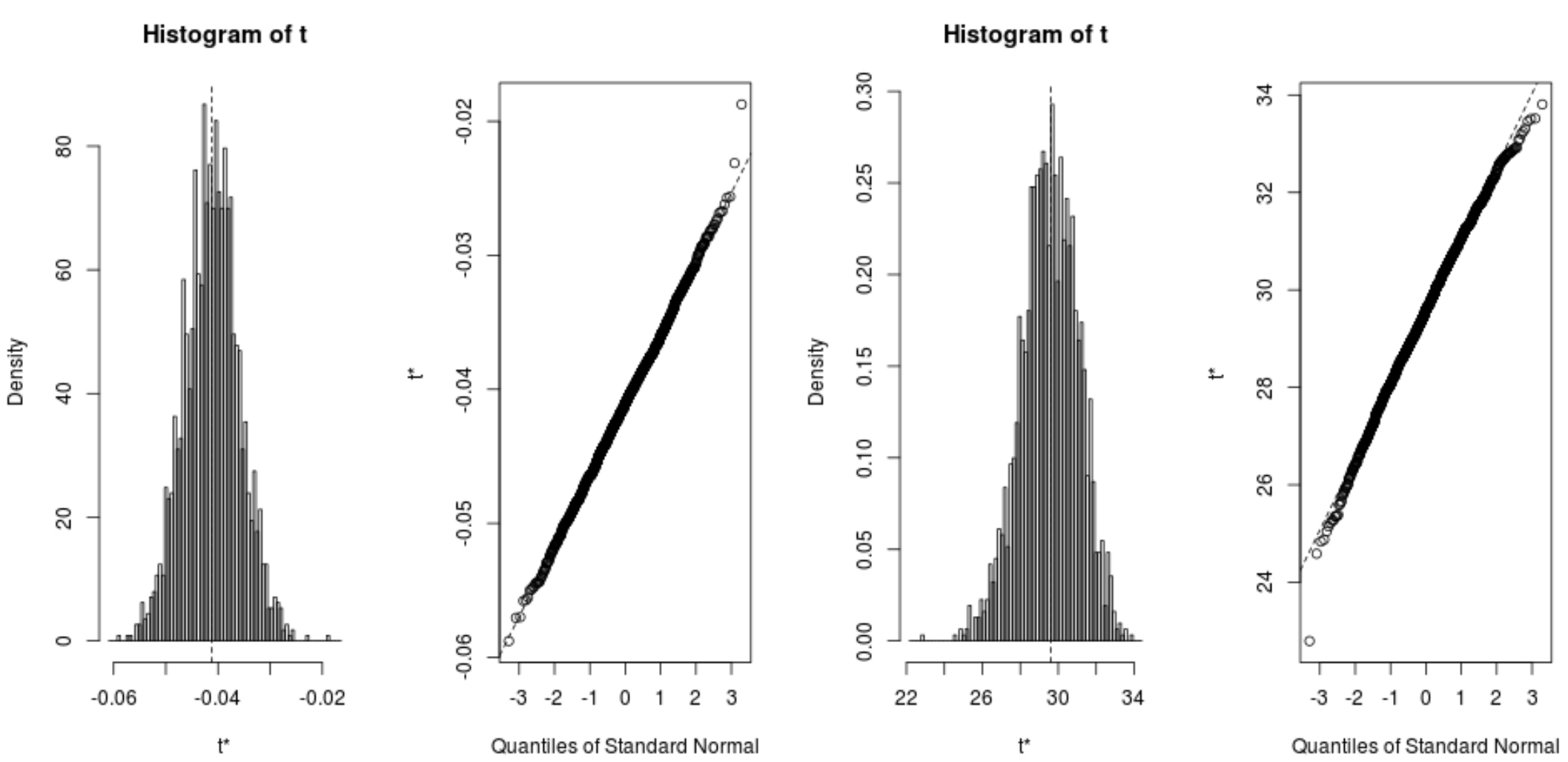
We can also use the following code to calculate the 95% confidence intervals for each coefficient:
#calculate adjusted bootstrap percentile (BCa) intervals boot.ci(reps, type="bca", index=1) #intercept of model boot.ci(reps, type="bca", index=2) #disp predictor variable CALL : boot.ci(boot.out = reps, type = "bca", index = 1) Intervals : Level BCa 95% (26.78, 32.66 ) Calculations and Intervals on Original Scale BOOTSTRAP CONFIDENCE INTERVAL CALCULATIONS Based on 2000 bootstrap replicates CALL : boot.ci(boot.out = reps, type = "bca", index = 2) Intervals : Level BCa 95% (-0.0520, -0.0312 ) Calculations and Intervals on Original Scale
From the output we can see that the 95% bootstrapped confidence intervals for the model coefficients are as follows:
- C.I. for intercept: (26.78, 32.66)
- C.I. for disp: (-.0520, -.0312)
How to Perform Simple Linear Regression in R
How to Perform Multiple Linear Regression in R
Introduction to Confidence Intervals
