Table of Contents
The lm() function in R can be used to extract residuals from a linear model. The residuals are the differences between the observed data and the predicted values from the linear model. The residuals can be extracted by using the resid() function on the output from the lm() function. This function will return the residuals for each data point in the linear model.
You can use the following syntax to extract the from the function in R:
fit$residuals
This example assumes that we used the lm() function to fit a linear regression model and named the results fit.
The following example shows how to use this syntax in practice.
Related:
Example: How to Extract Residuals from lm() in R
Suppose we have the following data frame in R that contains information about the minutes played, total fouls, and total points scored by 10 basketball players:
#create data frame df <- data.frame(minutes=c(5, 10, 13, 14, 20, 22, 26, 34, 38, 40), fouls=c(5, 5, 3, 4, 2, 1, 3, 2, 1, 1), points=c(6, 8, 8, 7, 14, 10, 22, 24, 28, 30)) #view data frame df minutes fouls points 1 5 5 6 2 10 5 8 3 13 3 8 4 14 4 7 5 20 2 14 6 22 1 10 7 26 3 22 8 34 2 24 9 38 1 28 10 40 1 30
Suppose we would like to fit the following multiple linear regression model:
points = β0 + β1(minutes) + β2(fouls)
We can use the lm() function to fit this regression model:
#fit multiple linear regression model
fit <- lm(points ~ minutes + fouls, data=df)
We can then type fit$residuals to extract the residuals of the model:
#extract residuals from model
fit$residuals
1 2 3 4 5 6 7
2.0888729 -0.7982137 0.6371041 -3.5240982 1.9789676 -1.7920822 1.9306786
8 9 10
-1.7048752 0.5692404 0.6144057
Since there were 10 total observations in our data frame, there are 10 residuals – one for each observation.
For example:
- The first observation has a residual value of 2.089.
- The second observation has a residual value of -0.798.
- The third observation has a residual value of 0.637.
We can then create a residual vs. fitted values plot if we’d like:
#store residuals in variable
res <- fit$residuals
#produce residual vs. fitted plot
plot(fitted(fit), res)
#add a horizontal line at 0
abline(0,0)
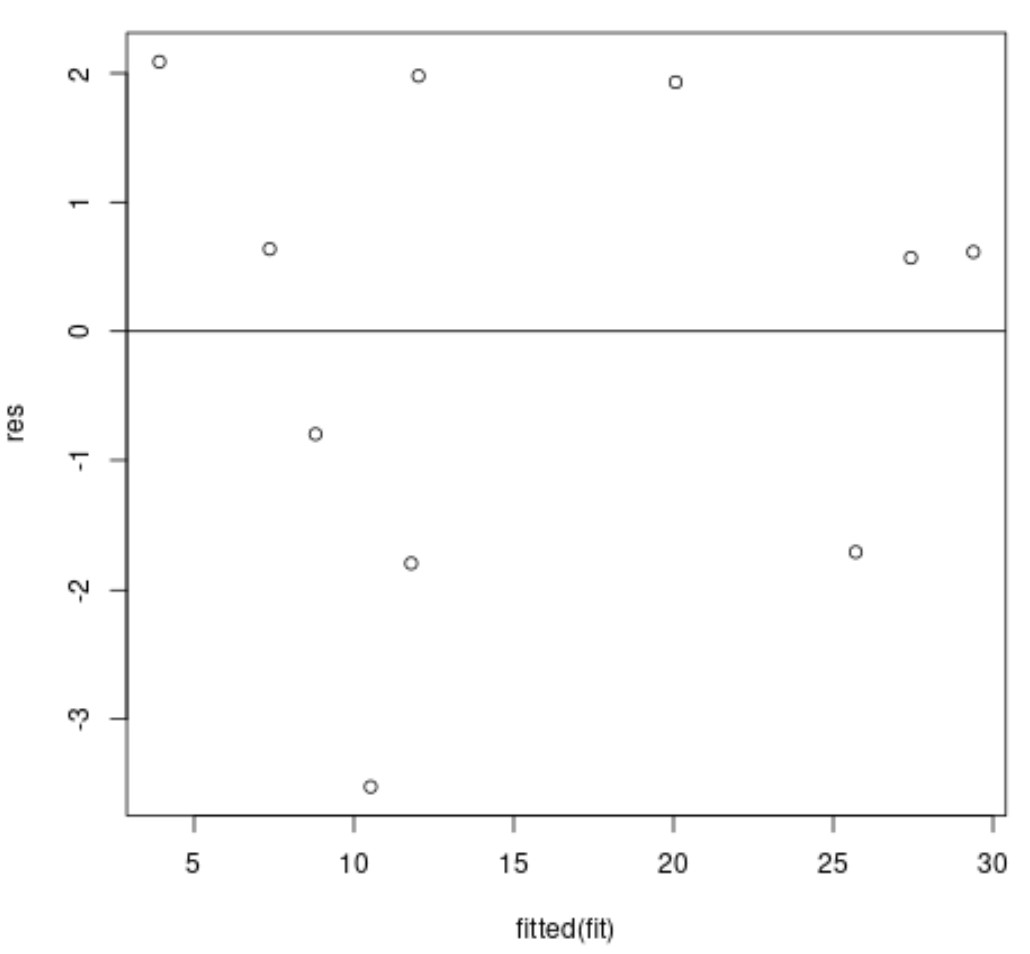
The x-axis displays the fitted values and the y-axis displays the residuals.
Ideally, the residuals should be randomly scattered about zero with no clear pattern to ensure that the is met.
In the residual plot above we can see that the residuals do seem to be randomly scatted about zero with no clear pattern, which means the assumption of homoscedasticity is likely met.
The following tutorials explain how to perform other common tasks in R:
