Table of Contents
LOESS regression is a non-parametric regression technique that uses locally weighted linear regression to fit a non-linear relationship between a response variable and one or more predictor variables. In R, it can be performed by using the “loess” function within the “stats” package. As an example, the following code could be used to fit a LOESS regression of the “mtcars” data set in R, where mpg is the response variable and wt is the predictor variable. loess(mpg ~ wt, data = mtcars)
LOESS regression, sometimes called local regression, is a method that uses local fitting to fit a regression model to a dataset.
The following step-by-step example shows how to perform LOESS regression in R.
Step 1: Create the Data
First, let’s create the following data frame in R:
#view DataFrame df <- data.frame(x=c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14), y=c(1, 4, 7, 13, 19, 24, 20, 15, 13, 11, 15, 18, 22, 27)) #view first six rows of data frame head(df) x y 1 1 1 2 2 4 3 3 7 4 4 13 5 5 19 6 6 24
Step 2: Fit Several LOESS Regression Models
We can use the loess() function to fit several LOESS regression models to this dataset, using various values for the span parameter:
#fit several LOESS regression models to dataset
loess50 <- loess(y ~ x, data=df, span=.5)
smooth50 <- predict(loess50)
loess75 <- loess(y ~ x, data=df, span=.75)
smooth75 <- predict(loess75)
loess90 <- loess(y ~ x, data=df, span=.9)
smooth90 <- predict(loess90)
#create scatterplot with each regression line overlaid
plot(df$x, df$y, pch=19, main='Loess Regression Models')
lines(smooth50, x=df$x, col='red')
lines(smooth75, x=df$x, col='purple')
lines(smooth90, x=df$x, col='blue')
legend('bottomright', legend=c('.5', '.75', '.9'),
col=c('red', 'purple', 'blue'), pch=19, title='Smoothing Span')
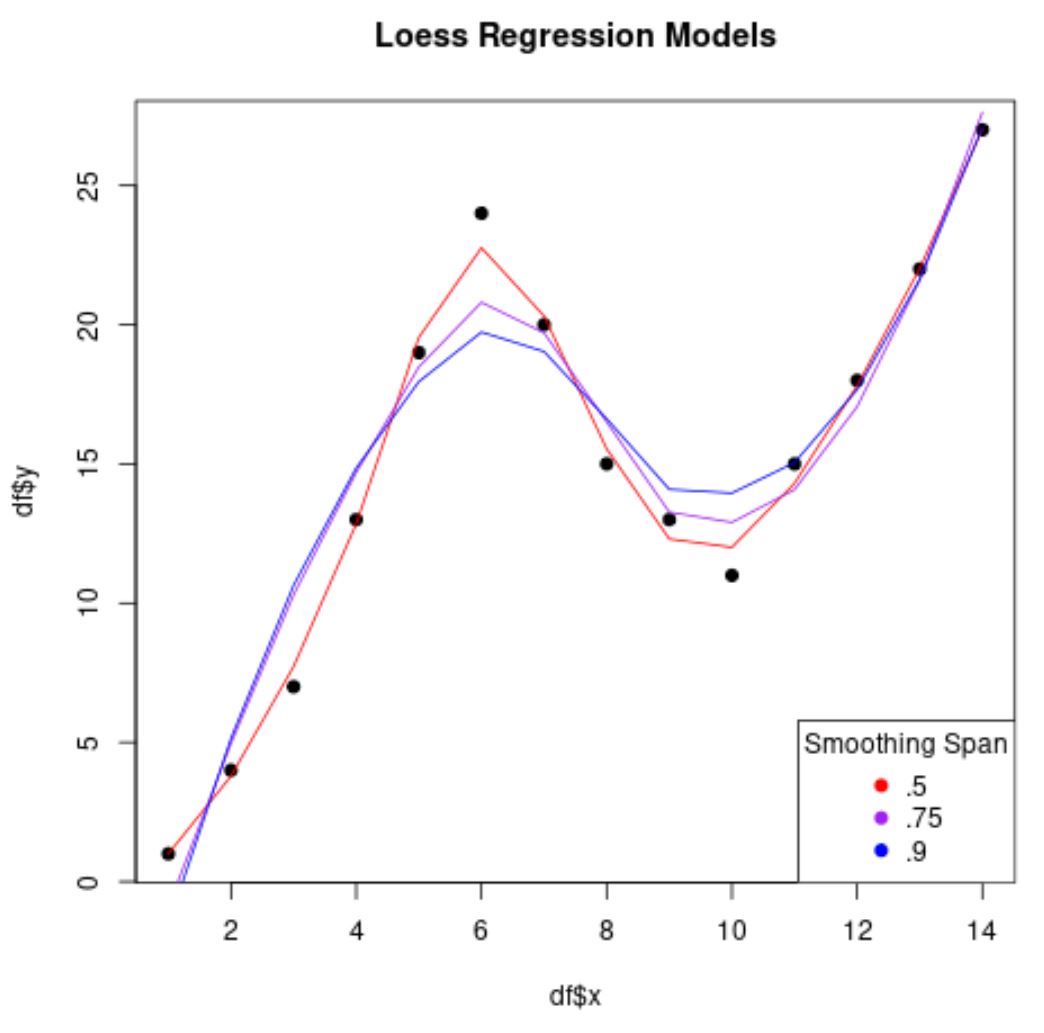
Notice that the lower the value that we use for span, the less “smooth” the regression model will be and the more the model will attempt to hug the data points.
Step 3: Use K-Fold Cross Validation to Find the Best Model
To find the optimal span value to use, we can perform using functions from the caret package:
library(caret)
#define k-fold cross validation method
ctrl <- trainControl(method = "cv", number = 5)
grid <- expand.grid(span = seq(0.5, 0.9, len = 5), degree = 1)
#perform cross-validation using smoothing spans ranging from 0.5 to 0.9
model <- train(y ~ x, data = df, method = "gamLoess", tuneGrid=grid, trControl = ctrl)
#print results of k-fold cross-validation
print(model)
14 samples
1 predictor
No pre-processing
Resampling: Cross-Validated (5 fold)
Summary of sample sizes: 12, 11, 11, 11, 11
Resampling results across tuning parameters:
span RMSE Rsquared MAE
0.5 10.148315 0.9570137 6.467066
0.6 7.854113 0.9350278 5.343473
0.7 6.113610 0.8150066 4.769545
0.8 17.814105 0.8202561 11.875943
0.9 26.705626 0.7384931 17.304833
Tuning parameter 'degree' was held constant at a value of 1
RMSE was used to select the optimal model using the smallest value.
The final values used for the model were span = 0.7 and degree = 1.
We can see that the value for span that produced the lowest value for the (RMSE) is 0.7.
Thus, for our final LOESS regression model we would choose to use a value of 0.7 for the span argument within the loess() function.
The following tutorials provide additional information about regression models in R:
