Table of Contents
The Cramer-Von Mises test in R is a non-parametric test for comparing two samples to determine whether they come from the same distribution. It can be performed using the cvm.test() function in R, which takes two numeric vectors as its arguments. An example of performing this test is to first create two vectors of data and then use the cvm.test() function to compare them. The output of the test will be the test statistic and the corresponding p-value, which can be used to determine if the two samples have a significant difference between them.
The Cramer-Von Mises test is used to determine whether or not a sample comes from a normal distribution.
This type of test is useful for determining whether or not a given dataset comes from a normal distribution, which is a used in many statistical tests including regression, ANOVA, t-tests, and many others.
We can easily perform a Cramer-Von Mises test using the cvm.test() function from the goftest package in R.
The following example shows how to use this function in practice.
Example 1: Cramer-Von Mises Test on Normal Data
The following code shows how to perform a Cramer-Von Mises test on a dataset with a sample size n=100:
library(goftest) #make this example reproducible set.seed(0) #create dataset of 100 random values generated from a normal distribution data <- rnorm(100) #perform Cramer-Von Mises test for normality cvm.test(data, 'pnorm') Cramer-von Mises test of goodness-of-fit Null hypothesis: Normal distribution Parameters assumed to be fixed data: data omega2 = 0.078666, p-value = 0.7007
The p-value of the test turns out to be 0.7007.
Since this value is not less than .05, we can assume the sample data comes from a population that is normally distributed.
This result shouldn’t be surprising since we generated the sample data using the rnorm() function, which generates random values from a .
Related: A Guide to dnorm, pnorm, qnorm, and rnorm in R
We can also produce a histogram to visually verify that the sample data is normally distributed:
hist(data, col='steelblue')
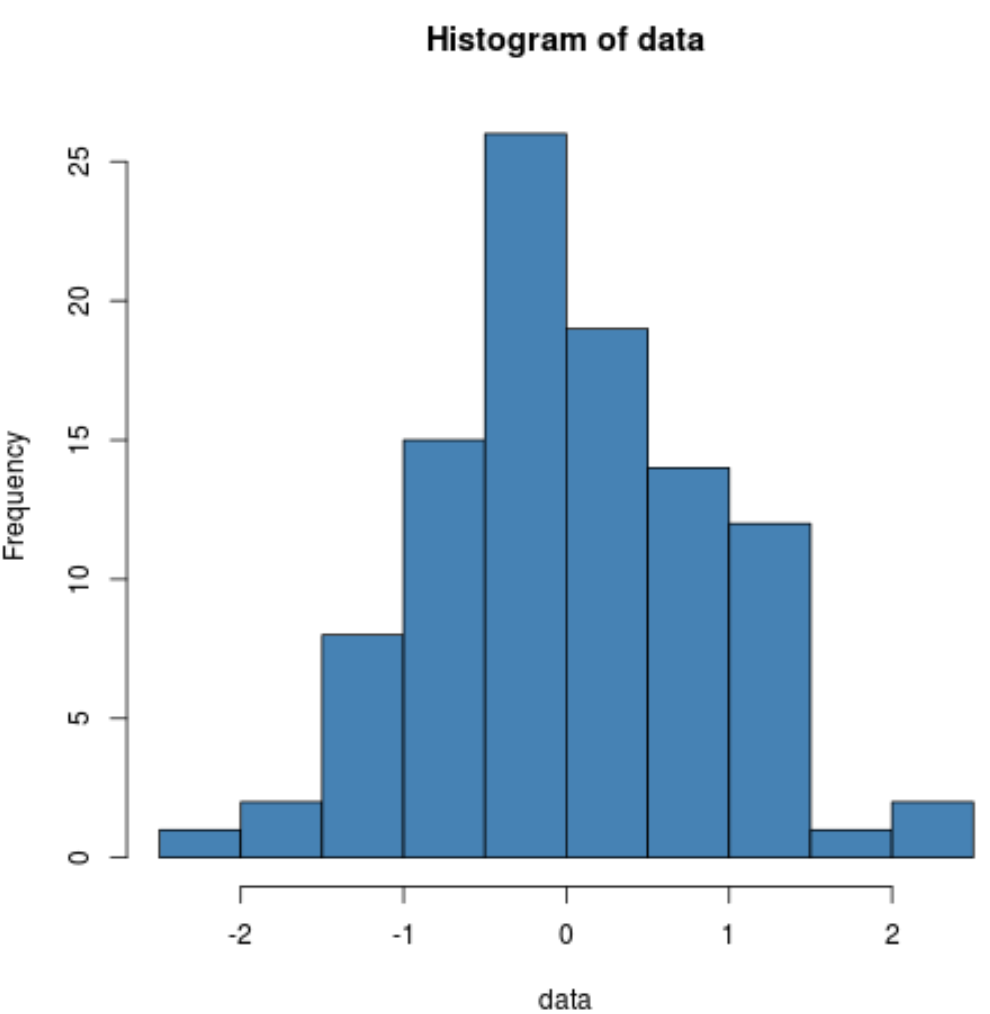
We can see that the distribution is fairly bell-shaped with one peak in the center of the distribution, which is typical of data that is normally distributed.
Example 2: Cramer-Von Mises Test on Non-Normal Data
The following code shows how to perform a Cramer-Von Mises test on a dataset with a sample size of 100 in which the values are randomly generated from a Poisson distribution:
library(goftest) #make this example reproducible set.seed(0) #create dataset of 100 random values generated from a Poisson distribution data <- rpois(n=100, lambda=3) #perform Cramer-Von Mises test for normality cvm.test(data, 'pnorm') Cramer-von Mises test of goodness-of-fit Null hypothesis: Normal distribution Parameters assumed to be fixed data: data omega2 = 27.96, p-value < 2.2e-16
The p-value of the test turns out to be extremely small.
Since this value is less than .05, we have sufficient evidence to say that the sample data does not come from a population that is normally distributed.
This result shouldn’t be surprising since we generated the sample data using the rpois() function, which generates random values from a Poisson distribution.
Related: A Guide to dpois, ppois, qpois, and rpois in R
We can also produce a histogram to visually see that the sample data is not normally distributed:
hist(data, col='coral2')
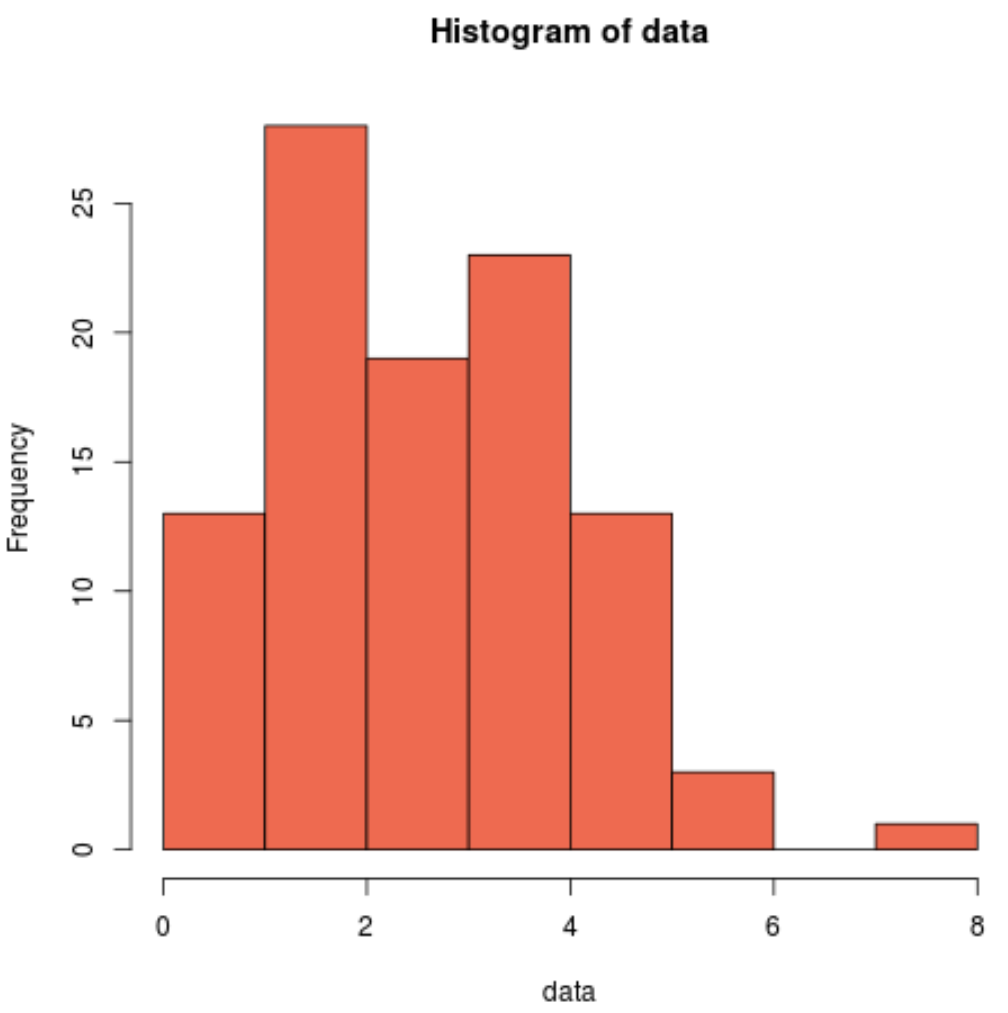
We can see that the distribution is and doesn’t have the typical “bell-shape” associated with a normal distribution.
Thus, our histogram matches the results of the Cramer-Von Mises test and confirms that our sample data does not come from a normal distribution.
What to Do with Non-Normal Data
If a given dataset is not normally distributed, we can often perform one of the following transformations to make it more normal:
1. Log Transformation: Transform the response variable from y to log(y).
2. Square Root Transformation: Transform the response variable from y to √y.
3. Cube Root Transformation: Transform the response variable from y to y1/3.
By performing these transformations, the response variable typically becomes closer to normally distributed.
Refer to this tutorial to see how to perform these transformations in practice.
The following tutorials explain how to perform other normality tests in R:
