Table of Contents
A MANOVA (Multivariate Analysis of Variance) analysis is a statistical technique used to analyze the relationship between multiple dependent variables and one or more independent variables. It is commonly used in research and data analysis to determine if there is a significant difference between groups on multiple variables.
To conduct a MANOVA analysis in R, you will need to first load the necessary packages, import your data, and make sure the data is in the correct format. Then, you will use the “manova” function to run the analysis, specifying the dependent and independent variables, and any other necessary parameters. The output of the analysis will provide information on the overall significance of the relationship between the variables and the specific effects of the independent variables on each dependent variable.
It is important to note that conducting a MANOVA analysis in R requires a solid understanding of statistical concepts and the R programming language. It is recommended to consult with a statistician or take a course on R before attempting to conduct a MANOVA analysis. Additionally, it is important to carefully interpret the results and consider potential confounding factors in your analysis.
Conduct a MANOVA in R
To understand the MANOVA, it first helps to understand the ANOVA.
An (analysis of variance) is used to determine whether or not there is a statistically significant difference between the means of three or more independent groups.
For example, suppose we want to know whether or not studying technique has an impact on exam scores for a class of students. We randomly split the class into three groups. Each group uses a different studying technique for one month to prepare for an exam. At the end of the month, all of the students take the same exam.
To find out if studying technique impacts exam scores, we can conduct a one-way ANOVA, which will tell us if if there is a statistically significant difference between the mean scores of the three groups.
In an ANOVA, we have one response variable. However, in a MANOVA (multivariate analysis of variance) we have multiple response variables.
For example, suppose we want to know how level of education (i.e. high school, associates degree, bachelors degrees, masters degree, etc.) impacts both annual income and amount of student loan debt. In this case, we have one factor (level of education) and two response variables (annual income and student loan debt), so we could conduct a one-way MANOVA.
Related:
How to Conduct a MANOVA in R
In the following example, we’ll illustrate how to conduct a one-way MANOVA in R using the built-in dataset iris, which contains information about the length and width of different measurements of flowers for three different species (“setosa”, “virginica”, “versicolor”):
#view first six rows of iris dataset
head(iris)
# Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#1 5.1 3.5 1.4 0.2 setosa
#2 4.9 3.0 1.4 0.2 setosa
#3 4.7 3.2 1.3 0.2 setosa
#4 4.6 3.1 1.5 0.2 setosa
#5 5.0 3.6 1.4 0.2 setosa
#6 5.4 3.9 1.7 0.4 setosa
Suppose we want to know if species has any effect on sepal length and sepal width. Using species as the independent variable, and sepal length and sepal width as the response variables, we can conduct a one-way MANOVA using the manova() function in R.
The manova() function uses the following syntax:
manova(cbind(rv1, rv2, …) ~ iv, data)
where:
- rv1, rv2: response variable 1, response variable 2, etc.
- iv: independent variable
- data: name of the data frame
#fit the MANOVA model model <- manova(cbind(Sepal.Length, Sepal.Width) ~ Species, data = iris) #view the results summary(model) # Df Pillai approx F num Df den Df Pr(>F) #Species 2 0.94531 65.878 4 294 < 2.2e-16 *** #Residuals 147 #--- #Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
From the output we can see that the F-statistic is 65.878 and the corresponding p-value is extremely small. This indicates that there is a statistically significant difference in sepal measurements based on species.
Technical Note: By default, manova() uses the Pillai test statistic. Since the distribution of this test statistic is complex, an approximate F value is also provided for easier interpretation.
In addition, it’s possible to specify “Roy”, “Hotelling-Lawley”, or “Wilks” as the test statistic to be used by using the following syntax: summary(model, test = ‘Wilks’)
To find out exactly how both sepal length and sepal width are affected by species, we can perform univariate ANOVAs using summary.aov() as shown in the following code:
summary.aov(model) # Response Sepal.Length : # Df Sum Sq Mean Sq F value Pr(>F) #Species 2 63.212 31.606 119.26 < 2.2e-16 *** #Residuals 147 38.956 0.265 #--- #Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 # Response Sepal.Width : # Df Sum Sq Mean Sq F value Pr(>F) #Species 2 11.345 5.6725 49.16 < 2.2e-16 *** #Residuals 147 16.962 0.1154 #--- #Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
We can see from the output that the p-values for both univariate ANOVAs are extremely small (<2.2e-16), which indicates that species has a statistically significant effect on both sepal width and sepal length.
Visualizing Group Means
It can also be helpful to visualize the group means for each level of our independent variable species to gain a better understanding of our results.
For example, we can use the gplots library and the plotmeans() function to visualize the mean sepal length by species:
#load gplots library library(gplots) #visualize mean sepal length by speciesplotmeans(iris$Sepal.Length ~ iris$Species)
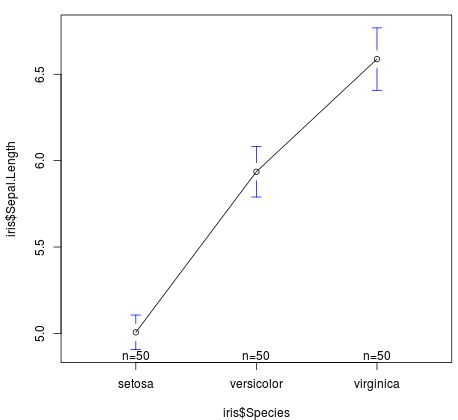
From the plot we can see that the mean sepal length varies quite a bit by species. This matches the results from our MANOVA, which told us that there was a statistically significant difference in sepal measurements based on species.
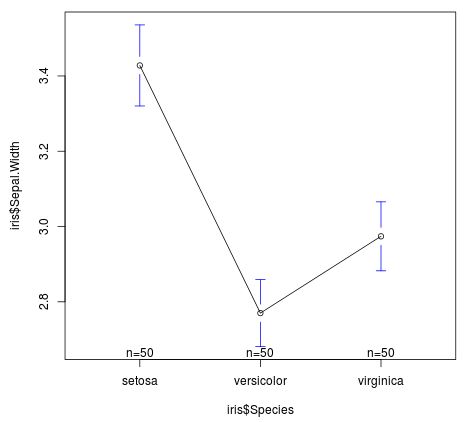
We can also visualize the mean sepal width by species:
plotmeans(iris$Sepal.Width ~ iris$Species)
View the full RDocumentation for the manova() function .
