Table of Contents
The Bonferroni correction is a method used to adjust the significance level of a hypothesis test when multiple tests are run simultaneously. To perform a Bonferroni correction in R, the user must use the p.adjust() function with the method argument set to “bonferroni” and the p-values of their hypothesis tests as the vector of p-values to be adjusted. This function will then return the adjusted p-values which can be used in place of the original p-values.
A one-way ANOVA is used to determine whether or not there is a statistically significant difference between the means of three or more independent groups.
If the overall p-value from the ANOVA table is less than some significance level, then we have sufficient evidence to say that at least one of the means of the groups is different from the others.
However, this doesn’t tell us which groups are different from each other. It simply tells us that not all of the group means are equal.
In order to find out exactly which groups are different from each other, we must conduct pairwise t-tests between each group while controlling for the family-wise error rate.
One of the most common ways to do so is to use Bonferroni’s correction when calculating the p-values for each of the pairwise t-tests.
This tutorial explains how to perform Bonferroni’s correction in R.
Example: Bonferroni’s Correction in R
Suppose a teacher wants to know whether or not three different studying techniques lead to different exam scores among students.
To test this, she randomly assigns 10 students to use each studying technique. After one week of using their assigned study technique, each student takes the same exam.
We can use the following steps in R to fit a one-way ANOVA and use Bonferroni’s correction to calculate pairwise differences between the exam scores of each group.
Step 1: Create the dataset.
The following code shows how to create a dataset that contains exam scores for all 30 students:
#create data frame data <- data.frame(technique = rep(c("tech1", "tech2", "tech3"), each = 10), score = c(76, 77, 77, 81, 82, 82, 83, 84, 85, 89, 81, 82, 83, 83, 83, 84, 87, 90, 92, 93, 77, 78, 79, 88, 89, 90, 91, 95, 95, 98)) #view first six rows of data frame head(data) technique score 1 tech1 76 2 tech1 77 3 tech1 77 4 tech1 81 5 tech1 82 6 tech1 82
Step 2: Visualize the exam scores for each group.
The following code shows how to produce boxplots to visualize the distribution of exam scores for each group:
boxplot(score ~ technique,
data = data,
main = "Exam Scores by Studying Technique",
xlab = "Studying Technique",
ylab = "Exam Scores",
col = "steelblue",
border = "black")
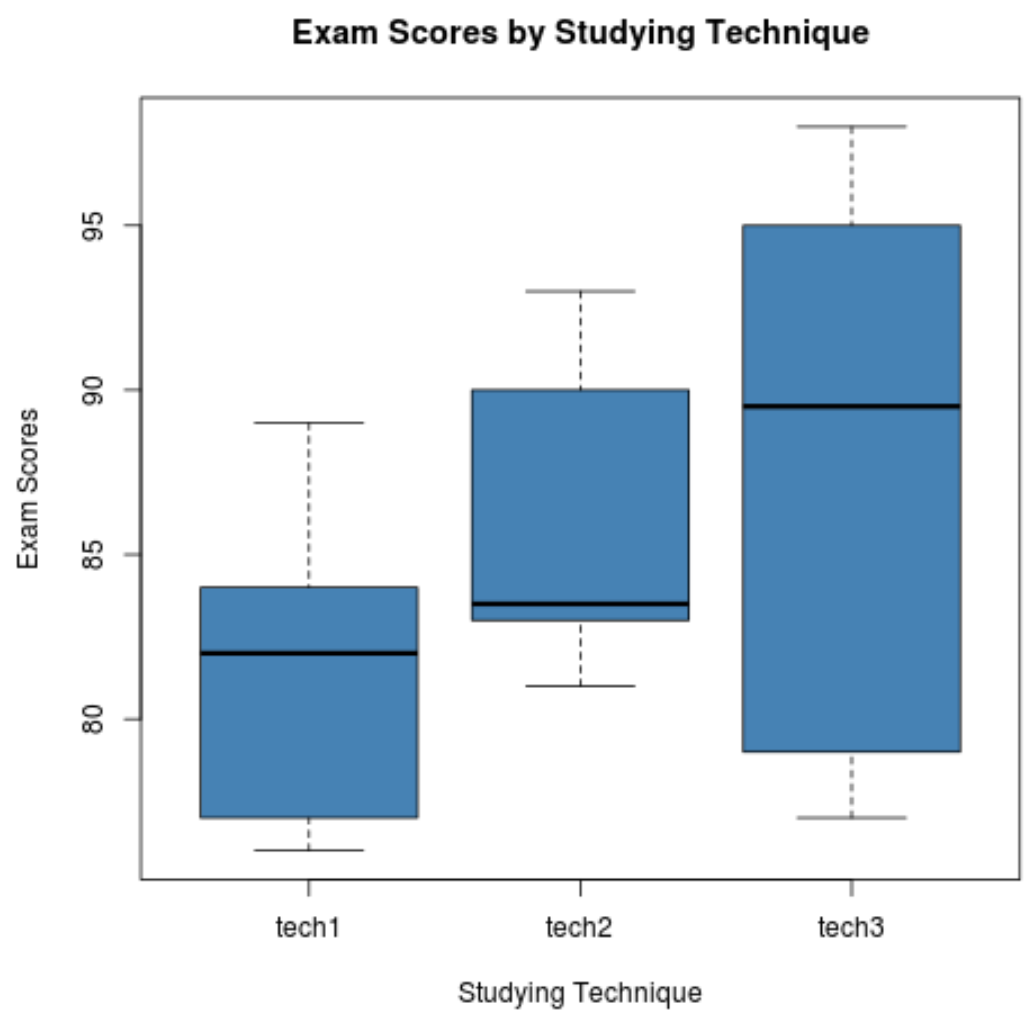
The following code shows how to perform a one-way ANOVA to test for differences among mean exam scores in each group:
#fit the one-way ANOVA model model <- aov(score ~ technique, data = data) #view model output summary(model) Df Sum Sq Mean Sq F value Pr(>F) technique 2 211.5 105.73 3.415 0.0476 * Residuals 27 836.0 30.96 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Since the overall p-value (0.0476) is less than .05, this is an indication that each group does not have the same average exam score.
Next, we will perform pairwise t-tests using Bonferroni’s correction for the p-values to calculate pairwise differences between the exam scores of each group.
Step 4: Perform pairwise t-tests.
To perform pairwise t-tests with Bonferroni’s correction in R we can use the pairwise.t.test() function, which uses the following syntax:
pairwise.t.test(x, g, p.adjust.method=”bonferroni”)
where:
- x: A numeric vector of response values
- g: A vector that specifies the group names (e.g. studying technique)
The following code shows how to use this function for our example:
#perform pairwise t-tests with Bonferroni's correction pairwise.t.test(data$score, data$technique, p.adjust.method="bonferroni") Pairwise comparisons using t tests with pooled SD data: data$score and data$technique tech1 tech2 tech2 0.309 - tech3 0.048 1.000 P value adjustment method: bonferroni
The way to interpret the output is as follows:
- The adjusted p-value for the mean difference in exam scores between technique 1 and technique 2 is .309.
- The adjusted p-value for the mean difference in exam scores between technique 1 and technique 3 is .048.
- The adjusted p-value for the mean difference in exam scores between technique 2 and technique 3 is 1.000.
Based on the output, we can see that the only significant difference is between technique 1 and technique 3.
An Introduction to the One-Way ANOVA
How to Conduct a One-Way ANOVA in R
How to Perform Tukey’s Test in R
How to Perform Dunnett’s Test in R
