Table of Contents
Cook’s Distance is a measure of the influence of a data point on a regression model, and can be used to identify outliers in the data. In Python, Cook’s Distance can be calculated by first fitting a linear regression model to the data, then calculating the residuals from the model and taking the sum of the squared residuals for each data point, and dividing by the sum of all squared residuals. This will give the Cook’s Distance for each data point, which can then be used to identify potential outliers.
Cook’s distance is used to identify influential in a regression model.
The formula for Cook’s distance is:
Di = (ri2 / p*MSE) * (hii / (1-hii)2)
where:
- ri is the ith residual
- p is the number of coefficients in the regression model
- MSE is the mean squared error
- hii is the ith leverage value
Essentially Cook’s distance measures how much all of the fitted values in the model change when the ith observation is deleted.
The larger the value for Cook’s distance, the more influential a given observation.
A general rule of thumb is that any observation with a Cook’s distance greater than 4/n (where n = total observations) is considered to be highly influential.
This tutorial provides a step-by-step example of how to calculate Cook’s distance for a given regression model in Python.
Step 1: Enter the Data
First, we’ll create a small dataset to work with in Python:
import pandas as pd #create dataset df = pd.DataFrame({'x': [8, 12, 12, 13, 14, 16, 17, 22, 24, 26, 29, 30], 'y': [41, 42, 39, 37, 35, 39, 45, 46, 39, 49, 55, 57]})
Step 2: Fit the Regression Model
Next, we’ll fit a :
import statsmodels.api as sm
#define response variable
y = df['y']
#define explanatory variable
x = df['x']
#add constant to predictor variables
x = sm.add_constant(x)
#fit linear regression model
model = sm.OLS(y, x).fit()
Step 3: Calculate Cook’s Distance
Next, we’ll calculate Cook’s distance for each observation in the model:
#suppress scientific notation
import numpy as np
np.set_printoptions(suppress=True)
#create instance of influence
influence = model.get_influence()
#obtain Cook's distance for each observation
cooks = influence.cooks_distance
#display Cook's distances
print(cooks)
(array([0.368, 0.061, 0.001, 0.028, 0.105, 0.022, 0.017, 0. , 0.343,
0. , 0.15 , 0.349]),
array([0.701, 0.941, 0.999, 0.973, 0.901, 0.979, 0.983, 1. , 0.718,
1. , 0.863, 0.713]))
By default, the cooks_distance() function displays an array of values for Cook’s distance for each observation followed by an array of corresponding p-values.
For example:
- Cook’s distance for observation #1: .368 (p-value: .701)
- Cook’s distance for observation #2: .061 (p-value: .941)
- Cook’s distance for observation #3: .001 (p-value: .999)
And so on.
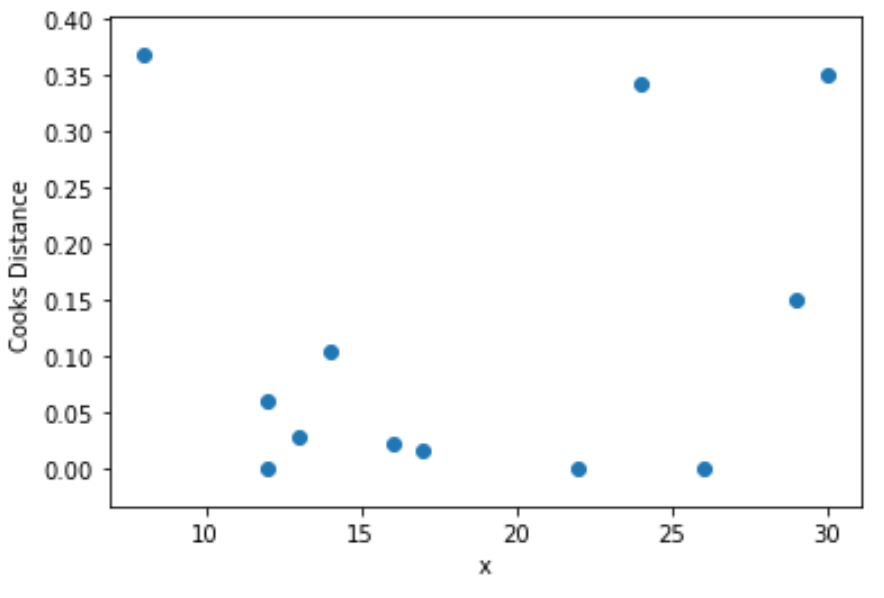
Step 4: Visualize Cook’s Distances
Lastly, we can create a scatterplot to visualize the values for the predictor variable vs. Cook’s distance for each observation:
import matplotlib.pyplot as plt
plt.scatter(df.x, cooks[0])
plt.xlabel('x')
plt.ylabel('Cooks Distance')
plt.show()
Closing Thoughts
It’s important to note that Cook’s Distance should be used as a way to identify potentially influential observations. Just because an observation is influential doesn’t necessarily mean that it should be deleted from the dataset.
First, you should verify that the observation isn’t a result of a data entry error or some other odd occurrence. If it turns out to be a legit value, you can then decide if it’s appropriate to delete it, leave it be, or simply replace it with an alternative value like the median.
