Table of Contents
The pheatmap() function in R can be used to create heatmaps from a given data matrix. This function clusters data in the matrix and displays it in a colored cluster heatmap. It also allows for some customization such as color scale, font size, clustering method, annotation, and number of columns and rows. The heatmap created can then be used to visualize the data and help identify patterns or correlations.
You can use the pheatmap() function from the pheatmap package in R to create highly customized heatmaps.
The following examples show how to use this function in practice with the following fake dataset:
#make this example reproducible set.seed(1) #create matrix with fake data values data = matrix(rnorm(100), 20, 5) data [1:10, seq(1, 5, 1)] = data [1:10, seq(1, 5, 1)] + 3 data [11:20, seq(2, 5, 1)] = data [11:20, seq(2, 5, 1)] + 2 data [15:20, seq(2, 5, 1)] = data [15:20, seq(2, 5, 1)] + 4 #add column names and row names colnames(data) = paste("T", 1:5, sep = "") rownames(data) = paste("Gene", 1:20, sep = "") #view matrx data T1 T2 T3 T4 T5 Gene1 2.37354619 3.918977 2.8354764 5.401618 2.431331 Gene2 3.18364332 3.782136 2.7466383 2.960760 2.864821 Gene3 2.16437139 3.074565 3.6969634 3.689739 4.178087 Gene4 4.59528080 1.010648 3.5566632 3.028002 1.476433 Gene5 3.32950777 3.619826 2.3112443 2.256727 3.593946 Gene6 2.17953162 2.943871 2.2925048 3.188792 3.332950 Gene7 3.48742905 2.844204 3.3645820 1.195041 4.063100 Gene8 3.73832471 1.529248 3.7685329 4.465555 2.695816 Gene9 3.57578135 2.521850 2.8876538 3.153253 3.370019 Gene10 2.69461161 3.417942 3.8811077 5.172612 3.267099 Gene11 1.51178117 3.358680 2.3981059 2.475510 1.457480 Gene12 0.38984324 1.897212 1.3879736 1.290054 3.207868 Gene13 -0.62124058 2.387672 2.3411197 2.610726 3.160403 Gene14 -2.21469989 1.946195 0.8706369 1.065902 2.700214 Gene15 1.12493092 4.622940 7.4330237 4.746367 7.586833 Gene16 -0.04493361 5.585005 7.9803999 6.291446 6.558486 Gene17 -0.01619026 5.605710 5.6327785 5.556708 4.723408 Gene18 0.94383621 5.940687 4.9558654 6.001105 5.426735 Gene19 0.82122120 7.100025 6.5697196 6.074341 4.775387 Gene20 0.59390132 6.763176 5.8649454 5.410479 5.526599
Example 1: Create Basic Heatmap
We can create a heatmap with the default settings in pheatmap to visualize all of the values in the matrix:
library(pheatmap)
#create basic heatmap
pheatmap(data)
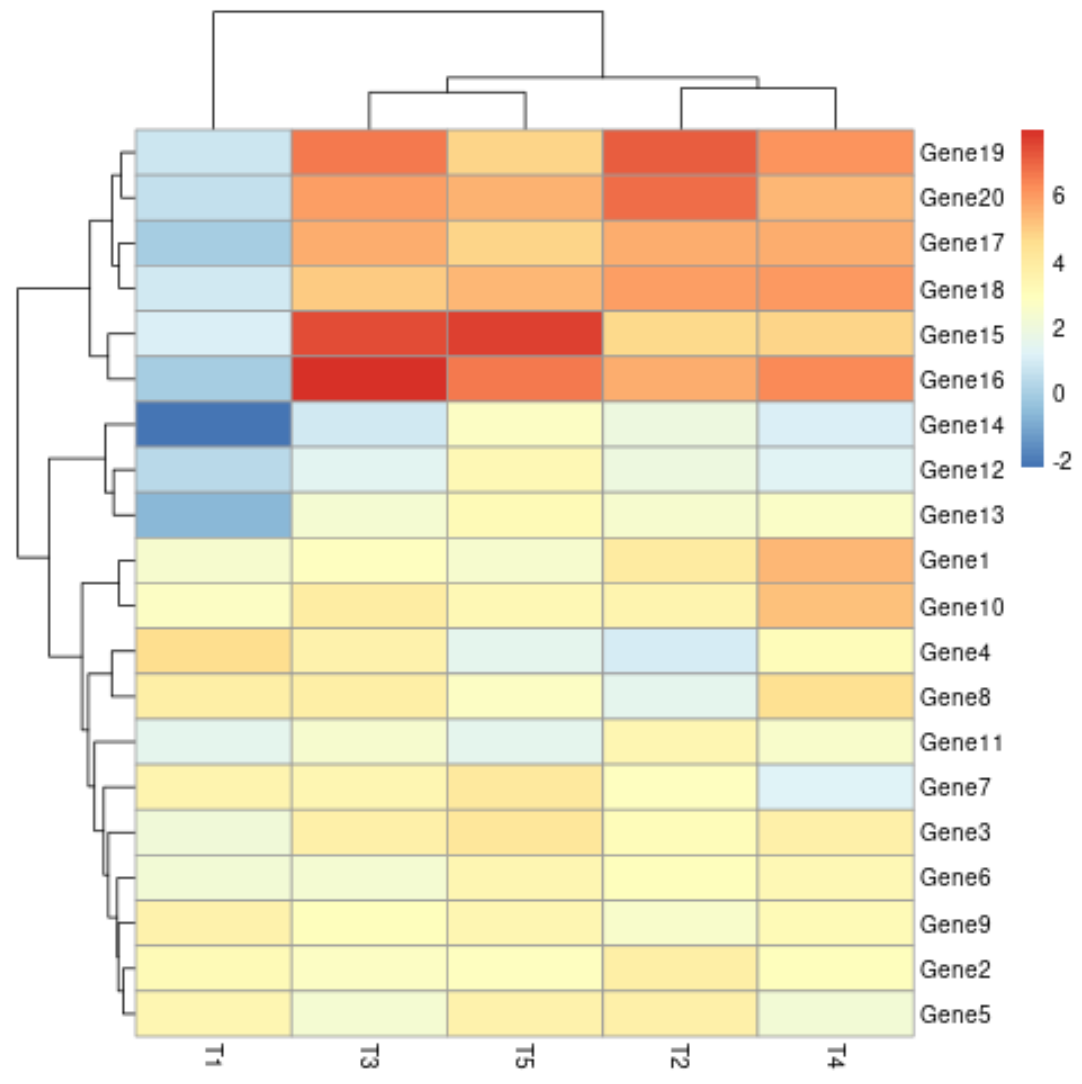
Example 2: Create Heatmap with Cell Labels
We can create use the arguments display_numbers and fontsize_number to display the numerical values in each cell of the heatmap with a specific font size:
library(pheatmap)
#create heatmap with numerical labels in cells
pheatmap(data, display_numbers=TRUE, fontsize_number=12)
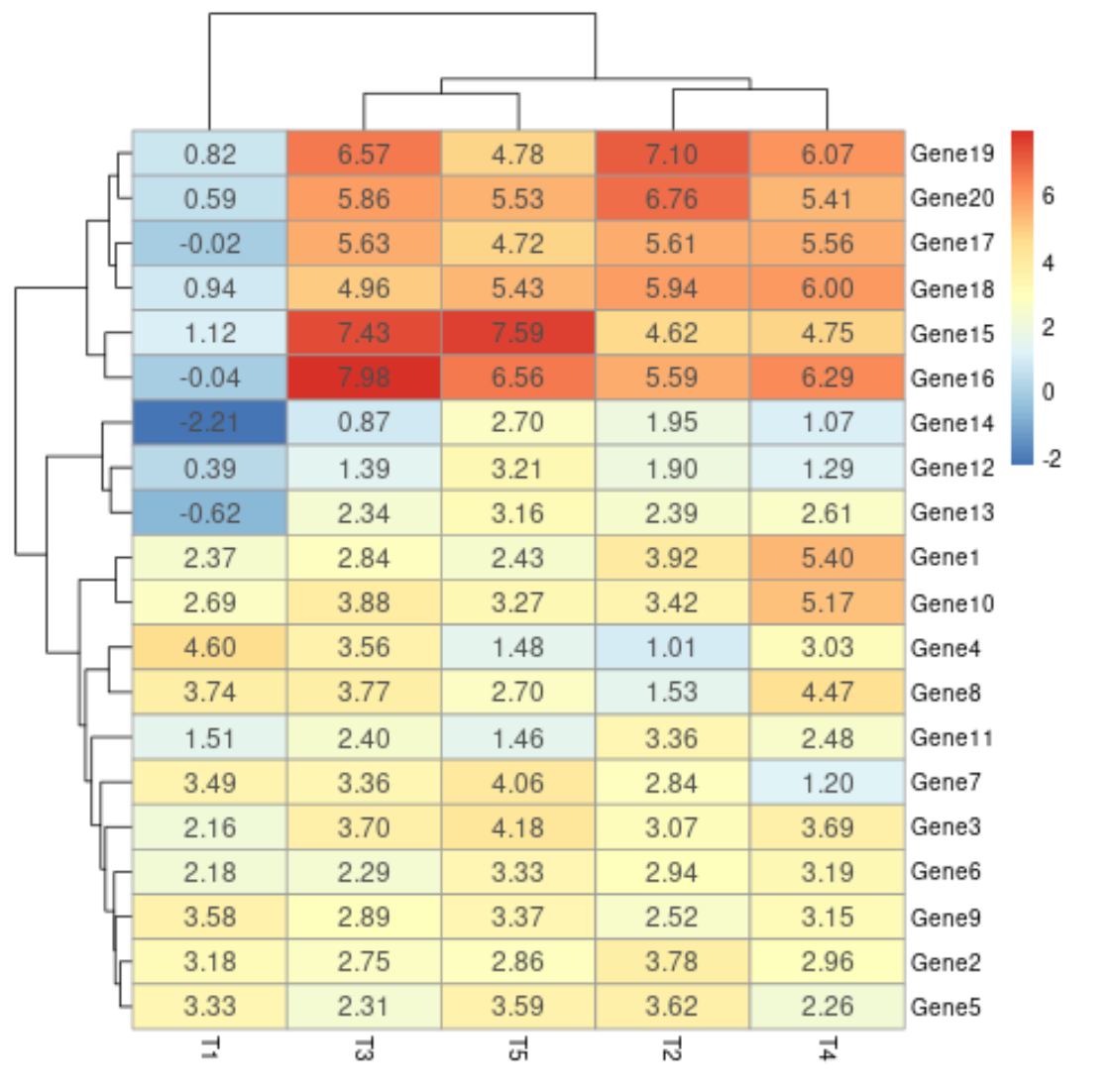
Note: The default value for fontsize_number is 8.
Example 3: Create Heatmap with Custom Colors
We can create use the colorRampPalette argument to specify the colors to use for the low, medium and high values in the heatmap as well:
library(pheatmap)
#create heatmap with custom colors
pheatmap(data, color=colorRampPalette(c("blue", "white", "red"))(20))
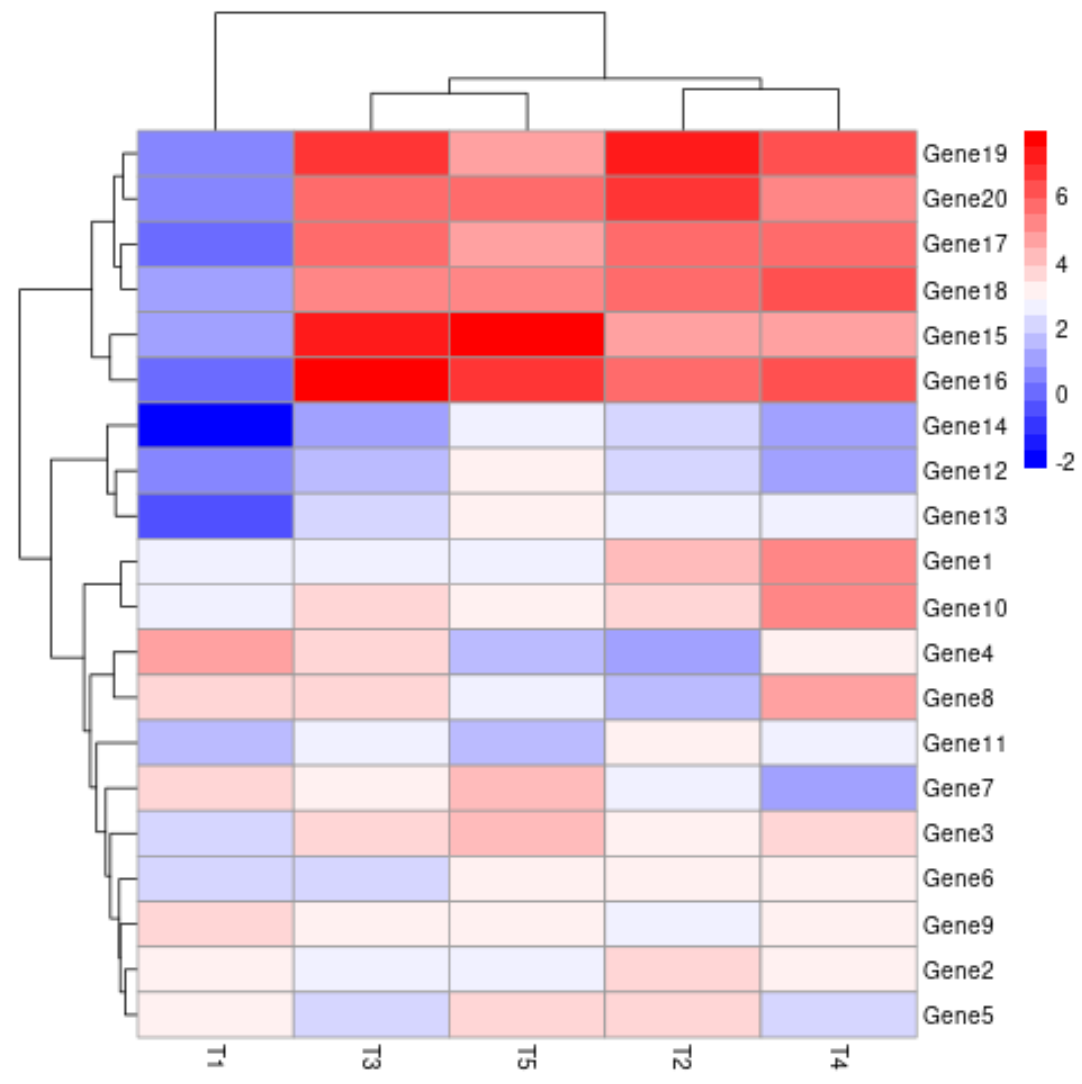
The low values are now shown in blue, the middle values are shown in white, and the high values are shown in red.
The following tutorials explain how to perform other common tasks in R:
